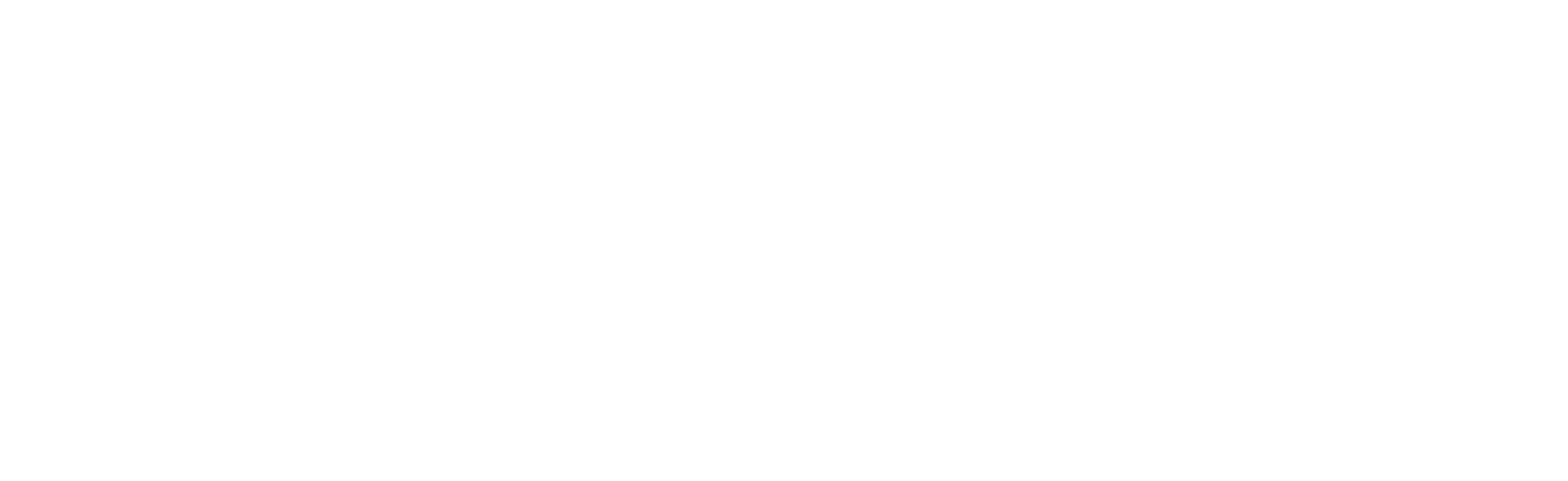![]()
Purpose:
The goal of this study is to better understand why rheumatic diseases happen, how they progress over time, and what makes each patient’s experience unique. By collecting and studying blood and other samples, along with basic medical information, our researchers hope to learn what’s happening at the cellular and genetic level. This can help us uncover patterns, identify early signs and symptoms, and possibly find better ways to diagnose, treat, and even prevent these diseases in the future.
In simple terms: Your participation helps researchers take a closer look at what’s going on in the body during autoimmune disease, and that knowledge can help improve care for future patients.
What We Collect:
Participants may be asked to donate blood (up to 80 mL per visit). These samples are stored securely and de-identified in our biorepository to support ongoing research in rheumatology and autoimmune diseases.
Meet The Coordinator:
Dr. Neela Moadab is the clinical research coordinator for the Rheumatic Disease Biorepository. She will be your main point of contact throughout the study, available to answer any questions, explain procedures, and help make your participation as smooth and comfortable as possible. Whether you are donating a sample or returning for a follow-up, Dr. Moadab will be there to guide and support you every step of the way.

Location:
University of Washington, Division of Rheumatology , Seattle, WA
Contact us: RHMRESEARCH@UW.EDU | (206) 616-5330
Recent Publications:
Expression of L1 retrotransposons in granulocytes from patients with active systemic lupus erythematosus
Date published:
10-May-23
DOI:
https://doi.org/10.1186/s13100-023-00293-7
Link:
https://mobilednajournal.biomedcentral.com/articles/10.1186/s13100-023-00293-7
First author:
Kennedy Ukadike, MD MS
Corresponding author:
Tomas Mustelin, MD PhD
The researchers were studying blood samples from patients with systemic lupus erythematosus (SLE). They made several key findings:
Protein Detection:
- They found a specific protein called ORF1p mainly in certain immune cells (neutrophils) of SLE patients
- Patients with more active disease had higher levels of this protein
- In patients with active disease, about 29% of their neutrophils contained this protein, compared to only about 2% in patients with inactive disease
Gene Activity:
- They identified specific genetic sequences (called L1Hs) that were producing this protein
- Each patient had a unique pattern of these active genes
- Patients with more active disease tended to have more of these genes turned on
Control Mechanisms:
- They found that some of the normal control mechanisms that usually keep these genes inactive were reduced in SLE patients
- This might explain why these genes become more active in SLE
Immune System Effects:
- The researchers found that these cells were producing interferon (an immune system signaling molecule)
- When they treated the cells with certain drugs (reverse transcriptase inhibitors), the production of interferon decreased
- This suggests that the activity of these genes might be contributing to the disease
Clinical Implications:
- The findings suggest that these genetic elements (L1Hs) might play a role in SLE
- The research points to the possibility that drugs targeting these elements might be helpful in treating SLE
- The researchers suggest that clinical trials with specific drugs (reverse transcriptase inhibitors) might be worth pursuing
This research helps understand how SLE works and suggests potential new ways to treat it, though more research would be needed to confirm these findings.
Expression of Envelope Protein Encoded by Endogenous Retrovirus K102 in Rheumatoid Arthritis Neutrophils
Date published:
17-May-23
DOI:
https://doi.org/10.3390/microorganisms11051310
Link:
https://www.mdpi.com/2076-2607/11/5/1310
First author:
Amanda Laine, MS
Corresponding author:
Tomas Mustelin, MD PhD
The causes of rheumatoid arthritis (RA) are not completely understood but are thought to be related to a chemical modification of proteins (citrullination), which triggers an immune response.
This study focuses on HERV-K, ancient viral genes in our DNA which usually stays dormant.
- These are leftover pieces of viruses that infected our ancestors millions of years ago and became part of our genetic code.
- Unlike most other ancient retroviruses in our genome that have been damaged beyond function, some HERV-K viruses remain nearly intact and can potentially produce viral proteins. These retroviruses are normally silenced in healthy people but can become active in certain conditions like early development, cancer, HIV infection, and possibly autoimmune diseases.
The researchers used advanced sequencing technology to identify which specific HERV-K retroviruses are more active in RA patients. Here’s what they discovered:
- In RA patients, some of these viral genes become active, particularly one called "HERV-K102." When this happens, it produces a protein called "Env" that appears on the surface of certain immune cells (specifically neutrophils, a type of white blood cells).
The researchers found that RA patients' immune systems create antibodies that attack this Env protein. This suggests that the activation of these ancient viral genes might be contributing to the autoimmune response in RA.
- Interestingly, they found that this HERV-K102 gene becomes more active in response to female hormones (estrogen and progesterone), which might help explain why RA is more common in women than men (4:1 ratio).
This study helps us better understand what might be triggering or contributing to rheumatoid arthritis. By identifying that these ancient viral genes become active in RA patients and produce proteins that the immune system attacks, researchers might be able to develop new treatments that target this specific aspect of the disease.
- This work also suggests that what we used to think of as "junk DNA" (old viral genes in our genome) might actually play important roles in certain diseases when they become inappropriately activated.
Multiple RNA-binding proteins associated with long interspersed element-1 encoded ORF1p are targeted by the autoimmune response in systemic lupus erythematosus
Date published:
17-Oct-23
DOI:
https://doi.org/10.46439/immunol.2.022
Link:
First author:
Kennedy Ukadike, MD MS
Corresponding author:
Tomas Mustelin, MD PhD
In systemic lupus erythematosus (SLE), patients produce antibodies that attack their own proteins (called autoantibodies). The researchers discovered that lupus patients have autoantibodies against several proteins that are normally found together in cells.
The main findings:
- They found autoantibodies against 9 different proteins that are usually found together in cell structures
- These autoantibodies were higher in patients with active lupus symptoms compared to those with inactive disease
- The levels of these autoantibodies were connected to inflammation markers and specific symptoms like joint pain
- Healthy people generally didn't have these autoantibodies
Why this matters:
This helps explain how lupus develops and progresses.
- The findings suggest that when cells die abnormally, they release these proteins in clusters, which then trigger the immune system to attack them. Understanding this process could help in the development of better, more targeted treatments for lupus.
- The researchers also found that these autoantibodies tend to appear together - if a patient had antibodies against one of these proteins, they were likely to have antibodies against the others too.
In conclusion, this study helps us better understand how lupus attacks the body and why certain proteins become targets of the immune system in lupus patients. This knowledge could potentially lead to better ways to diagnose and treat the disease.
Prediction of alternative pre‑mRNA splicing outcomes
Date published:
15-Nov-23
DOI:
https://doi.org/10.1038/s41598-023-47348-6
Link:
https://www.nature.com/articles/s41598-023-47348-6
First and corresponding author:
Rayan Najjar, MD MS
Scientists developed a new computational tool called RiboSplitter that helps understand how genes can create different versions of proteins through a process called alternative splicing.
The Tool:
- RiboSplitter predicts how different versions of genes will be translated into proteins
- It can recognize new, previously unknown versions of genes
- It creates helpful visualizations to show how changes in gene splicing affect the final protein
- It's more efficient than previous tools because it filters out irrelevant data
The research team tested the efficacy of RiboSplitter in studying blood cells (specifically B cells) from patients with Sjögren's syndrome. Here's what they found:
- They identified 1,367 significant differences in how genes were spliced between healthy people and Sjögren's patients
- The tool successfully predicted how these changes would affect proteins in about 94% of cases
- About 10% of the changes found were completely new (never seen before)
- They discovered an interesting change in a gene called caspase 8, which is involved in cell death. In Sjögren's patients, the cells made more of the complete version of this protein, which could lead to increased cell death
A unique advancement is that RiboSplitter can:
- Work with both known and newly discovered gene versions
- Show exactly how changes in gene splicing affect the final protein structure
- Process data more efficiently than previous tools
- Create clear visualizations to help scientists understand the results
This tool is particularly valuable because it helps scientists understand not just that genes are being spliced differently in disease, but what those changes mean for the proteins they produce, which is crucial for understanding diseases and developing treatments.
Autoimmunity: the neoantigen hypothesis
Date published:
26-Jun-24
DOI:
10.3389/fimmu.2024.1432985
Link:
https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2024.1432985/full
First and corresponding author:
Tomas Mustelin, MD PhD
The paper challenges a long-held belief about the causes and development of autoimmune diseases.
Traditional View Being Challenged:
Scientists have long believed that autoimmune diseases are the result of our immune system "losing tolerance,” meaning it suddenly starts attacking our own body's healthy tissues by mistake.
New Perspective:
The authors propose a different explanation: Instead of the immune system malfunctioning, they suggest that autoimmune diseases occur because new, unfamiliar versions of proteins (called "neoantigens") appear in our body. The immune system isn't broken – it's actually doing its job by responding to these modified proteins it's never seen before.
They support this new theory with several key points:
- Different But Predictable: Autoimmune diseases have distinct patterns (affecting specific organs, being more common in certain genders or age groups). This suggests they're not random immune system failures.
- Modified Proteins: In many autoimmune diseases, normal proteins get chemically modified in ways that make them look foreign to the immune system. For example, in rheumatoid arthritis, normal proteins get modified through a process called citrullination.
- Treatment Patterns: Different autoimmune diseases respond to different treatments, suggesting they're caused by different types of modified proteins rather than a general immune system malfunction.
Practical Implications: If this theory is correct, it could lead to better treatments. Instead of just suppressing the immune system (current approach), future treatments could prevent these protein modifications from happening in the first place, potentially stopping the disease at its source.
This new perspective represents a significant shift in how we understand and treat autoimmune diseases.
Subcellular location of L1 retrotransposon-encoded ORF1p, reverse transcription products, and DNA sensors in lupus granulocytes
Date published:
27-Jun-24
DOI:
10.1186/s13100-024-00324-x
Link:
https://mobilednajournal.biomedcentral.com/articles/10.1186/s13100-024-00324-x#citeas
First author:
Fatemeh Moadab, MD
Corresponding author:
Tomas Mustelin, MD PhD
This study focuses on a key discovery about Systemic Lupus Erythematosus (SLE).
The Main Finding
Researchers found that neutrophils (a type of white blood cell) from lupus patients contain unusual protein clusters that include:
- ORF1p - a protein encoded by LINE-1 retrotransposons (ancient virus-like DNA sequences in our genome)
- Ro60 - a well-known lupus autoantigen (a protein that triggers autoimmune responses)
- MOV10 - a protein that normally helps defend against viruses
- Special DNA-RNA hybrid molecule
- ZBP1 - a sensor that detects unusual nucleic acids
What This Means
These protein clusters form "granules" that look like viruses to the immune system. When neutrophils die through inflammatory processes (like NETosis* or pyroptosis**), these granules escape into the bloodstream where they:
- Trigger the production an inflammatory signal (called interferon)
- Stimulate the immune system to make antibodies against the proteins in these granules
Clinical Correlation
Patients with more active lupus had:
- More neutrophils containing these granules
- Higher levels of antibodies against the proteins in these granules
- More evidence of neutrophil cell death
Potential Treatment Implications
The researchers found that drugs that inhibit reverse transcriptase (the enzyme that creates DNA-RNA hybrids) could prevent the formation of these inflammatory molecules. This suggests a potential new treatment approach for lupus.
Why This Matters
This study provides insight into the pathogenesis of lupus - showing how transposable elements or "jumping genes,” like LINE-1 retrotransposons, might contribute to autoimmunity when they become active in certain immune cells. The process creates a feedback loop where inflammation leads to cell death, which releases more inflammatory signals, perpetuating the disease.
*NETosis: the process by with neutrophils expel their nuclear DNA, referred to as nuclear extracellular traps (NETs). This happens more readily in neutrophils of SLE patients, compared to healthy donor neutrophils.
**pyroptosis: a type of programmed cell death that may contribute to lupus disease activity
“Escape of ORF1p granules from SLE neutrophils during NETosis.
A., ORF1p (red) in 6 or 7 SLE neutrophils undergoing NETosis. A normal neutrophil also appears in the bottom left panel. The approximate location of the neutrophil before NETosis is indicated with a dotted circle. Externalized, free or DNA-associated ORF1p granules are indicated with white arrows. B., ORF1p (red) remains intracellular in neutrophils undergoing apoptosis* as indicated by activated caspase-3** (green). C., ORF1p (red) in a neutrophil undergoing pyroptosis as indicated by cell ballooning and the absence of activated caspase-3 (green). At least one ORF1p granule appears to have escaped (white arrow). The shown images were taken with 100 X magnification.”
*Apoptosis: a type of programmed cell death
**Caspase-3: an enzyme that triggers apoptosis
Evidence of membranolytic targeting and intracellular citrullination in neutrophils isolated from patients with rheumatoid arthritis
Date published:
5-Jul-24
DOI:
https://doi.org/10.1038/s41598-024-66516-w
Link:
https://www.nature.com/articles/s41598-024-66516-w
First author:
Fatemeh Moadab, MD
Corresponding author:
Tomas Mustelin, MD PhD
In this study, researchers found evidence that immune cells are attacking white blood cells (specifically neutrophils) in patients with rheumatoid arthritis by creating tiny holes in them. This attack leads to chemical changes inside these white blood cells that may trigger or worsen the disease.
Background Context:
- RA is an autoimmune disease where the body's immune system attacks its own joints
- People with RA often have special antibodies (called anti-citrullinated protein antibodies, ACPA) that recognize proteins that have been chemically modified through a process called citrullination
Scientists didn't understand exactly how or why this modification occurs in RA.
Findings:
- The study team found that some white blood cells collected from RA patients had small holes in their surface made by a protein called perforin.
- These holes allow calcium to rush into the cells, triggering chemical changes (citrullination) inside.
- Healthy people's white blood cells didn't have these holes
- In a severe form of RA called Felty's syndrome, where patients have very low white blood cell counts, they found extremely high levels of ACPA
Why It's Important:
This research suggests a new understanding of how RA might develop–rather than the immune system randomly attacking joints, there may be a specific process where immune cells target white blood cells, leading to chemical changes that trigger the disease.
Potential Impact: This discovery could lead to new treatments that target this specific process rather than just suppressing the entire immune system, which is the current approach.
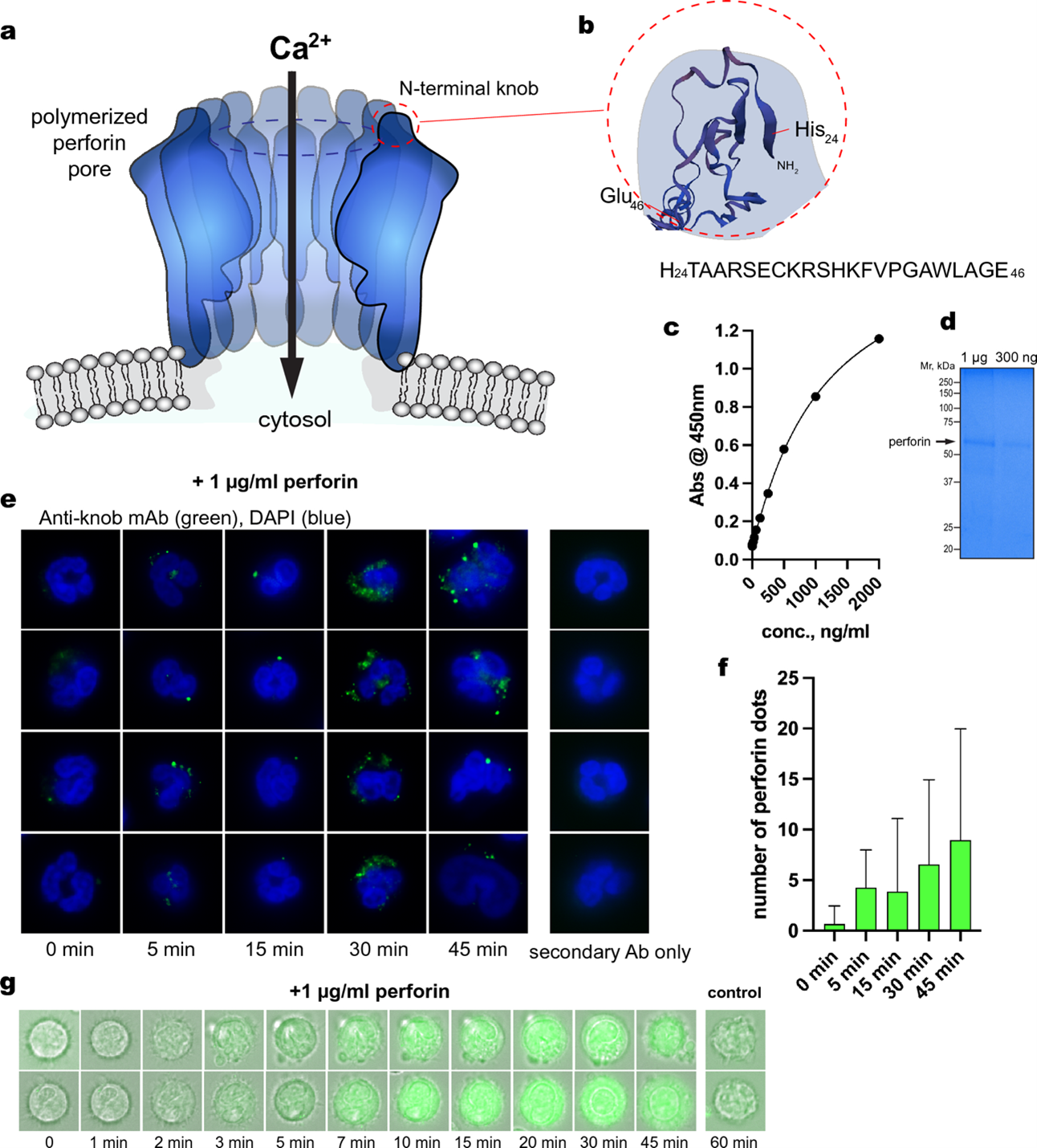
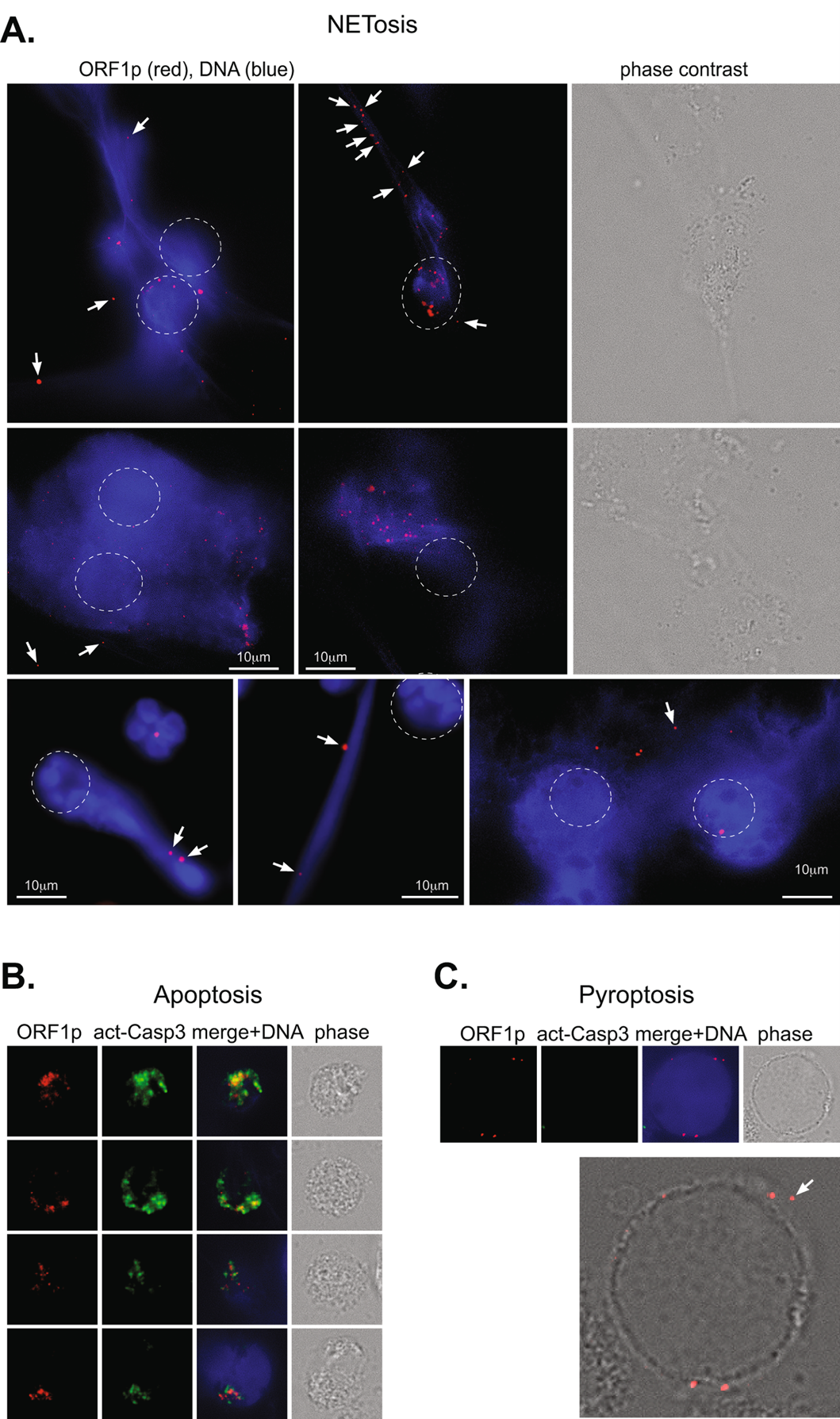
Figure 1. Generation and validation of the knob mAb (anti-perforinN). (a) The structure of the pore made in the cell membrane of RA neutrophils. The pore is comprised of perforin proteins and allows calcium (Ca2+) to enter the cell. (b) The N-terminal knob is the part of perforin that can be recognized by an antibody that the study team developed, called “anti-knob mAb 4G10”. (c) The results of a lab test (called an ELISA) showing that the anti-knob mAb 4G10 can, in fact, detect the N-terminal knob. (d) The result of a lab test (called Coomassie Blue staining) that help the lab team visualize the perforin protein. (e) Images of neutrophils treated with perforin for the listed time. The cells were immunoflourescently stained to show the anti-knob mAb 4G10 (green) and the cells’ DNA (blue). (f) The number of detected perforin dots (indicated by green dots in panel e) over time. (g) Images of 2 neutrophils treated with perforin and a fluorescent calcium indicator (brighter green indicates more calcium inside the cell).
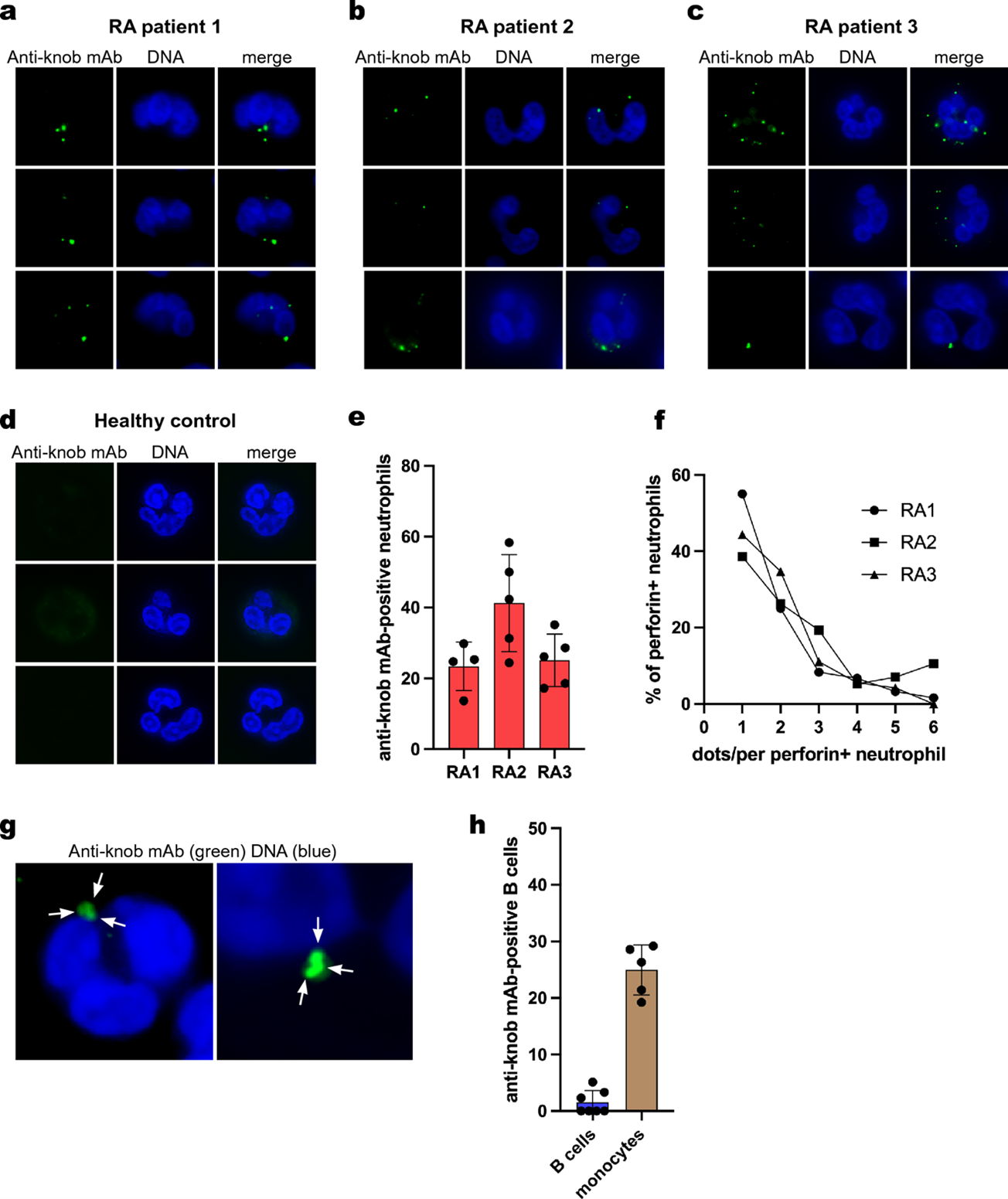
Figure 2. Poly-perforin pores on freshly isolated untreated RA neutrophils. (a), (b), (c) Images of 3 neutrophils from blood samples of 3 RA patients. The cells were stained to show the anti-knob mAb (green dots), which indicates the presence of perforin pores, and the cells’ DNA (blue). (d) Images of similarly stained neutrophils from a healthy donor, showing no perforin pores. (e) Graph showing the percentage of neutrophils with perforin pores in the same 3 RA patients as images in a, b, and c. (f) Graph showing the number of bright perforin dots on each of the perforin-positive neutrophils from the same three RA patients. (g) Close-up images of clusters of green dots. (h) Graph showing the number of perforin pores observed in RA B cells and monocytes (other types of white blood cells).